

Structural Characterization of RNA and Detection of RNA-ligand Binding Using Microfluidic Modulation Spectroscopy
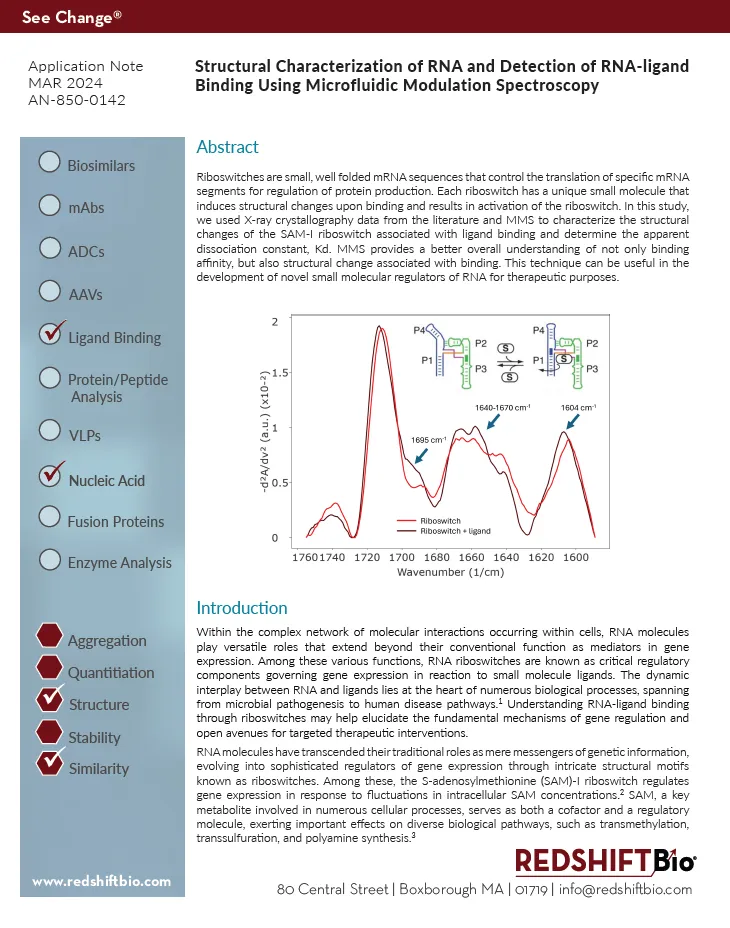
Riboswitches are small, well folded mRNA sequences that control the translation of specific mRNA segments for regulation of protein production. Each riboswitch has a unique small molecule that induces structural changes upon binding and results in activation of the riboswitch. In this study, we used X-ray crystallography data from the literature and MMS to characterize the structural changes of the SAM-I riboswitch associated with ligand binding and determine the apparent dissociation constant, Kd.
Microfluidic Modulation Spectroscopy is a proven method for accurate and sensitive characterization of proteins and nucleic acids and is being rapidly accepted as an essential technique for elucidating m RNA structure. In this application, MMS provides a better overall understanding of not only binding affinity, but also structural change associated with binding. This technique can be useful in the development of novel small molecular regulators of RNA for RNA therapeutic development.
Want to hear more from the study authors? Check out our recent webinar that features Arrakis Therapeutics and how they utilized MMS technology to gain insights into RNA structure change due to ligand binding!
Be sure to visit us at Formulation and Delivery UK/RNA Therapeutics & Delivery April 25-26, 2024 to learn more about MMS and its ability to accurately characterize RNA for therapeutic development!
Please complete the form to download the full app note.

