Ligand Binding Analysis and Protein Stabilization with MMS
Understanding Ligand Binding in Protein Therapeutics
Small molecules can be used to modify the activity, specificity, conformation, and stability of proteins. Traditionally, ligand binding affinity is quantitated using the dissociation constant, KD, due to the transient nature of the interactions. This quantifies what concentration of ligand is required for half of the binding sites to be occupied. Tightly binding ligands, i.e. ones with low KD values, are typically preferred because they will be more potent. Subsequent characterization would include activity assays to determine more specifically how the ligand affects the protein.
Challenges in Ligand Binding Characterization
One of the challenges with characterizing protein-ligand binding is the innate sensitivity required to observe a conformational change due to ligand binding in complex buffers.
How MMS Enhances Ligand Binding Analysis
The Aurora TX provides secondary structural information of the protein-ligand complex without the need of a tag or dye. This information adds to the overall picture of ligand binding strength and protein activity, offering insights into the potential binding mechanism. For example, if the ligand increases the protein activity because it alters the structure, Microfluidic Modulation Spectroscopy (MMS) would be able to quantitate those changes in the secondary structure.
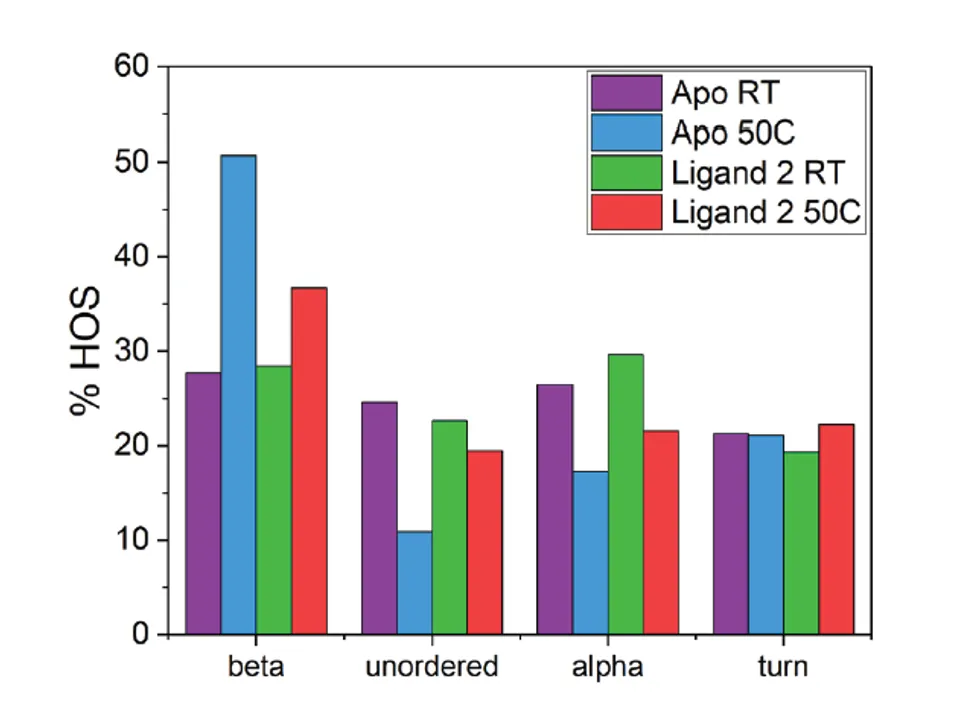
Example: Stabilizing Proteins with Ligand Binding
Another example is shown below, where the protein stability is greatly increased when bound to a ligand. In this example, the “Apo” sample is unbound and heating at 50°C causes aggregation and drastically increases the beta-sheet content compared to the room temperature (RT) sample. However, in the presence of “Ligand 2,” the same heat-stress causes a much less dramatic increase in aggregation and beta-sheet formation. This is an example of an accelerated stability study in which ligand binding provides protection against aggregation without causing major changes to the native structure at room temperature.